 Author
Author  Correspondence author
Correspondence author
Animal Molecular Breeding, 2024, Vol. 14, No. 2
Received: 27 Jan., 2024 Accepted: 05 Mar., 2024 Published: 16 Mar., 2024
Pork is a significant component of global meat consumption, and carcass traits directly impact the economic value of meat. Quantitative genetics provides essential tools and methods for understanding and improving these traits. This study explores the application of quantitative genetics in improving pig carcass traits, ex-amining heritability, genetic correlations of carcass traits, and the genetic architecture of economically im-portant carcass traits in pigs. Additionally, this study compares the effectiveness of traditional breeding methods, marker-assisted selection (MAS), and genomic selection (GS) in improving carcass traits, and pre-sents case studies demonstrating the practical application of these methods. The findings indicate that breed-ing strategies based on genetic information, such as MAS and GS, significantly enhance the efficiency of se-lecting for carcass traits. Case studies further validate the successful application of these strategies in re-al-world pig breeding programs and highlight areas for future research and improvement. By deeply under-standing the principles of quantitative genetics and their role in the selection of pig carcass traits, this study provides a theoretical foundation and practical guidance for future breeding programs. The integration of ad-vanced molecular marker technologies and genetic analysis methods can effectively improve breeding effi-ciency and accuracy, promoting the sustainable development of the pork industry.
1 Introduction
Carcass traits in pigs, such as muscle area, fat content, and meat quality, are critical factors in the swine industry due to their direct impact on economic returns and consumer satisfaction. These traits influence the yield and quality of pork products, which are essential for meeting market demands and ensuring profitability for producers. For instance, traits like backfat thickness, intramuscular fat content, and muscle pH are closely monitored as they affect both the visual appeal and the eating quality of pork (Miar et al., 2014a; Miar et al., 2014b). The integration of carcass and meat quality traits into breeding objectives has become increasingly important to enhance the overall value of pork products (Miar et al., 2014).
Quantitative genetics is a branch of genetics that deals with the inheritance of traits that are determined by multiple genes, often influenced by environmental factors. This field focuses on the statistical analysis of phenotypic variation to understand the genetic architecture of complex traits. Techniques such as genome-wide association studies (GWAS), quantitative trait locus (QTL) mapping, and heritability estimation are commonly used to identify genetic factors associated with economically important traits in livestock (Duarte et al., 2017; Falker-Gieske et al., 2019; Zhang et al., 2019). These methods allow researchers to dissect the genetic basis of traits and provide insights into the genetic correlations between different traits (Milan et al., 2002; Edwards et al., 2008).
In animal breeding, quantitative genetics plays a pivotal role in improving desirable traits through selective breeding programs. By understanding the genetic parameters, such as heritability and genetic correlations, breeders can make informed decisions to enhance traits like growth rate, carcass composition, and meat quality (Miar et al., 2014a; Soares et al., 2022). The application of quantitative genetics in pig breeding has led to significant advancements in the selection for traits that improve both production efficiency and product quality. For example, the identification of QTLs and significant SNPs associated with carcass and meat quality traits has facilitated the development of marker-assisted selection strategies, accelerating genetic improvement in pig populations (Čepica et al., 2013; Duarte et al., 2017).
This study evaluates the application of quantitative genetics in improving carcass traits in pigs. It summarizes current knowledge on the genetic basis of pig carcass traits, reviews the methodologies used in quantitative genetics to study these traits, assesses the effectiveness of quantitative genetics in enhancing carcass traits through breeding programs, identifies gaps in existing research, and suggests future directions for improving carcass traits using quantitative genetics, with the aim of providing a comprehensive understanding of how to optimize pig carcass traits through quantitative genetics, offering theoretical support for the development of the swine industry.
2 Fundamentals of Quantitative Genetics
2.1 Basic principles of quantitative genetics
Quantitative genetics is the study of traits that are influenced by multiple genes and environmental factors. These traits, known as quantitative or complex traits, exhibit continuous variation and are typically measured on a numerical scale. The fundamental principles of quantitative genetics involve understanding how genetic variation contributes to phenotypic variation within a population. This is achieved through the study of quantitative trait loci (QTL), which are regions of the genome that are associated with variation in a quantitative trait. For instance, in pigs, numerous QTL have been identified for traits such as growth, meat quality, and carcass composition, highlighting the polygenic nature of these economically important traits (Duarte et al., 2017; Falker-Gieske et al., 2019; Velez-Irizary et al., 2019).
The identification and analysis of QTL involve genome-wide association studies (GWAS) and other genetic mapping techniques. These methods allow researchers to pinpoint specific genetic variants that contribute to trait variation. For example, a study on pigs used a low-coverage whole-genome sequencing strategy to identify 14 QTLs associated with various agricultural traits, demonstrating the complex genetic architecture underlying these traits (Yang et al., 2021). Additionally, the use of structural equation models (SEQM) can help in modeling causal relationships between multiple variables, further elucidating the genetic basis of complex traits (Peñagaricano et al., 2015).
2.2 Heritability and its role in carcass trait selection
Heritability is a key concept in quantitative genetics, representing the proportion of phenotypic variation in a population that is attributable to genetic variation. High heritability indicates that a significant portion of the variation in a trait is due to genetic differences among individuals, making it a crucial factor in selective breeding programs. For carcass traits in pigs, heritability estimates can guide breeders in selecting animals with desirable genetic profiles to improve traits such as backfat thickness, loin weight, and meat quality (Polasik et al., 2018; Falker-Gieske et al., 2019).
The role of heritability in carcass trait selection is exemplified by studies that have identified specific genetic markers associated with these traits. For instance, the MYH7 single nucleotide polymorphism (SNP) has been linked to growth and carcass traits in pigs, with significant associations observed for traits like backfat thickness and loin eye area (Polasik et al., 2018). By understanding the heritability of these traits, breeders can make informed decisions to enhance the genetic potential of their herds, ultimately leading to improved carcass quality and economic gains.
2.3 Genetic correlation between different carcass traits
Genetic correlation refers to the extent to which different traits share common genetic determinants. In the context of carcass traits in pigs, understanding genetic correlations is essential for simultaneous improvement of multiple traits. Positive genetic correlations indicate that selection for one trait will result in a correlated response in another trait, while negative correlations suggest that improving one trait may adversely affect another. For example, a study on pigs revealed significant genetic correlations between various carcass traits, such as backfat thickness and meat percentage (Figure 1), highlighting the interconnected nature of these traits (Polasik et al., 2018; Falker-Gieske et al., 2019).
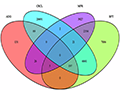 Figure 1 Analysis of Consistency and Inconsistency of Genetic Variations Among Different Traits (Adapted from Falker-Gieske et al., 2019) Image Caption: The Venn diagram illustrates the consistency and inconsistency of genetic variations among four traits: average daily gain (ADG), backfat thickness (BFT), meat-to-fat ratio (MFR), and carcass length (CRCL); The intersecting areas represent the number of shared genetic variations among these traits, while the non-overlapping areas indicate the number of unique variations specific to each trait (Adapted from Falker-Gieske et al., 2019). |
Falker-Gieske et al. (2019) found that carcass length (CRCL) exhibited a degree of genetic overlap with other traits such as MFR and BFT, suggesting a potential correlation between these traits at the genetic level. This correlation is significant for breeding strategies, as selecting for one trait (e.g., carcass length) could indirectly improve other highly correlated traits (e.g., backfat thickness or meat-to-fat ratio). However, Figure 1 also reveals unique genetic variations specific to each trait, indicating that these traits are independent in certain aspects and require targeted selection during breeding. Therefore, understanding the genetic correlations between these traits can help develop more effective breeding programs that achieve simultaneous improvement of target traits.
The identification of genetic correlations can also inform breeding strategies to avoid undesirable trade-offs. For instance, a study using epistatic QTL analysis found that certain genomic regions exhibited both positive and negative dominance effects on different carcass traits, such as entire belly weight and entire ham weight (Duthie et al., 2010). By considering these genetic correlations, breeders can develop more effective selection programs that optimize the overall genetic improvement of carcass traits.
2.4 Genetic architecture of economically important carcass traits in pigs
The genetic architecture of economically important carcass traits in pigs is complex, involving multiple genes and their interactions. Advances in sequencing technology and genome-wide association studies have significantly enhanced our understanding of this genetic complexity. For example, a meta-analysis of GWAS data identified key transcription factors and gene networks associated with meat quality and carcass traits, such as SOX5 and NKX2-5, which play crucial roles in adipose tissue metabolism and skeletal muscle development (Duarte et al., 2017).
Furthermore, the identification of novel genetic networks and epistatic interactions has provided deeper insights into the regulation of carcass traits. A study on beef cattle, which can be extrapolated to pigs, demonstrated that carcass traits rarely share genetic networks with eating quality and fatty acid composition traits, suggesting that marker-assisted selection for one category of traits would not interfere with the improvement of another (Jiang et al., 2009). This highlights the importance of understanding the genetic architecture to develop targeted breeding strategies that maximize the economic value of carcass traits in pigs.
3 Breeding Strategies for Improving Carcass Traits
3.1 Traditional breeding methods
Traditional breeding methods for improving carcass traits in pigs have relied heavily on phenotypic selection. This approach involves selecting animals based on observable traits such as growth rate, feed efficiency, and carcass quality. The primary advantage of this method is its simplicity and direct application, as it does not require advanced genetic knowledge or technology. However, the effectiveness of phenotypic selection is often limited by the heritability of the traits in question and the generation interval, which can slow down genetic progress (Biermann et al., 2015).
Despite its limitations, traditional breeding has been successful in achieving moderate improvements in carcass traits over time. For instance, the use of performance testing schemes that focus on phenotyped selection candidates has been shown to increase genetic gain for meat quality traits in local pig breeds (Biermann et al., 2015). However, the advent of molecular genetics and the identification of genetic markers have paved the way for more sophisticated breeding strategies that can potentially accelerate genetic improvement.
3.2 Marker-assisted selection (MAS)
Marker-assisted selection (MAS) integrates molecular genetics with traditional breeding methods by using genetic markers linked to desirable traits. This approach allows for the selection of animals based on their genetic potential rather than solely on phenotypic traits. MAS has proven effective for traits controlled by a few genes with large effects, such as certain qualitative traits (Budhlakoti et al., 2022). However, its application to quantitative traits, which are influenced by many genes with small effects, has been more challenging.
The efficiency of MAS in improving quantitative traits can be limited by the detectability of associations between marker loci and quantitative trait loci (QTL), as well as sampling errors in estimating the weighting coefficients in the selection index. Despite these challenges, MAS has been shown to increase selection efficiency when combined with phenotypic data. For example, a breeding strategy that includes both phenotypic and genetic marker information has resulted in a 20% increase in accuracy and selection response for meat quality traits in pigs (Biermann et al., 2015).
3.3 Genomic selection (GS)
Genomic selection (GS) represents a significant advancement over MAS by utilizing genome-wide markers to predict the breeding values of individuals. Unlike MAS, which focuses on a limited number of markers, GS incorporates all available marker information into the prediction model, thereby capturing the effects of numerous small-effect QTL (Heffner et al., 2009). This comprehensive approach allows for more accurate predictions of genetic potential and can substantially accelerate the breeding cycle (Heffner et al., 2010).
GS has been widely adopted in animal breeding programs due to its potential to improve selection accuracy, reduce the need for extensive phenotyping, and increase genetic gains (Budhlakoti et al., 2022). For instance, in pig breeding, GS has been shown to outperform MAS in terms of prediction accuracy and selection differentials (Arruda et al., 2016). The use of high-density SNP genotyping and advanced statistical models has further enhanced the effectiveness of GS, making it a powerful tool for improving complex traits such as carcass quality (Meuwissen et al., 2016; Merrick et al., 2022).
3.4 Comparison of different breeding strategies
When comparing traditional breeding methods, MAS, and GS, it is evident that each strategy has its strengths and limitations. Traditional breeding methods are straightforward and cost-effective but may be slow in achieving genetic progress due to the reliance on phenotypic selection and longer generation intervals (Biermann et al., 2015). MAS offers a more targeted approach by using genetic markers, but its effectiveness is limited for complex traits controlled by many genes (Budhlakoti et al., 2022).
In contrast, GS provides a more comprehensive and accurate method for predicting breeding values by incorporating genome-wide marker information. This approach has been shown to significantly accelerate genetic gains and improve selection accuracy compared to both traditional breeding and MAS (Heffner et al., 2009; Heffner et al., 2010; Arruda et al., 2016). However, the implementation of GS requires substantial investment in genotyping and computational resources, which may be a barrier for some breeding programs (Meuwissen et al., 2016; Merrick et al., 2022).
Overall, the choice of breeding strategy depends on the specific goals of the breeding program, the traits of interest, and the available resources. While traditional methods and MAS can still play a role in certain contexts, GS represents the most advanced and effective approach for improving complex traits such as carcass quality in pigs.
4 Case Study: Application of Quantitative Genetics in Pig Breeding Programs
4.1 Description of the breeding program
The breeding program under consideration involves a comprehensive approach to improving carcass traits in pigs through the application of quantitative genetics. This program integrates various genetic and phenotypic data to enhance economically important traits such as backfat thickness, loin depth, and carcass daily gain. For instance, in a study involving crossbred pigs, a reference population was used to evaluate the predictive ability of different models for carcass traits, with data collected from over 130 000 animals (Bergamaschi et al., 2019). Another breeding program focused on a three-generation experimental cross between Meishan and Large White pig breeds, analyzing 15 different carcass composition traits (Milan et al., 2002).
These breeding programs typically involve large-scale phenotypic measurements and genotyping efforts. For example, in the Duroc x Pietrain resource population, 510 F2 animals were genotyped for 124 microsatellite markers, and various carcass and meat quality traits were evaluated (Edwards et al., 2008). Similarly, a study on Duroc pigs estimated genetic parameters for 39 traits, including growth, conventional carcass traits, and novel carcass traits, using data from 2,583 purebred Duroc gilts (Willson et al., 2020).
4.2 Implementation of quantitative genetic methods
Quantitative genetic methods are implemented in these breeding programs through genome-wide association studies (GWAS), quantitative trait locus (QTL) mapping, and the estimation of genetic parameters. For instance, a GWAS conducted on pooled F2 designs led to the identification of numerous significantly associated variant clusters for traits such as average daily gain, backfat thickness, and carcass length (Falker-Gieske et al., 2019). In another study, QTL mapping in a Landrace pig population identified QTL for carcass weight, cutlet weight, and backfat thickness, among other traits (Vidal et al., 2005).
The use of genetic markers and heritability estimates is also crucial. In a study on the endangered German pig breed 'Bunte Bentheimer,' genetic markers at the ryanodine receptor 1 (RYR1) locus were used alongside phenotypic data to design breeding strategies for meat quality improvement (Biermann et al., 2015). Additionally, heritability estimates for various traits, such as meat lightness, loin pH, and marbling, were found to be moderate to high, indicating that these traits can be genetically improved if included in selection schemes (Willson et al., 2019).
4.3 Outcomes and improvements in carcass traits
The application of quantitative genetics in these breeding programs has led to significant improvements in carcass traits. For example, the pooling of four large F2 designs in a GWAS study resulted in the discovery of more than 32 million variants, including 8 million previously unreported ones, and the identification of new candidate genes such as BMP2 (Falker-Gieske et al., 2019). In another study, the inclusion of records from different finishing flows in the training set increased the prediction accuracy of carcass traits by approximately 6% (Bergamaschi et al., 2019).
Moreover, the identification of significant QTL for traits like backfat thickness, loin depth, and carcass daily gain has facilitated the fine mapping of genes controlling these traits, enabling their incorporation into marker-assisted selection programs (Edwards et al., 2008). The use of genetic parameters in Duroc pigs has also shown that genetic progress can be achieved for a wide range of traits, including novel carcass traits, without undesirable impacts on growth rate and carcass leanness (Willson et al., 2020).
4.4 Lessons learned and future perspectives
One of the key lessons learned from these breeding programs is the importance of integrating both phenotypic and genotypic data to achieve accurate predictions and significant genetic gains. The use of high-density genotyping and advanced statistical models has proven effective in identifying genetic markers and QTL associated with economically important traits (Bergamaschi et al., 2019; Falker-Gieske et al., 2019). Additionally, the inclusion of genetic marker information, such as the RYR1 locus, has been shown to increase the accuracy and selection response in breeding strategies (Biermann et al., 2015).
Looking forward, future perspectives include the continued refinement of genetic models and the incorporation of new technologies such as whole-genome sequencing and gene editing. The development of more precise and efficient selection indexes that include a broader range of traits, such as meat quality and novel carcass traits, will be essential for meeting the growing demand for high-quality pork (Duarte et al., 2017; Willson et al., 2020). Furthermore, the validation and functional characterization of identified QTL and candidate genes will be crucial for translating genetic findings into practical breeding applications (Vidal et al., 2005).
5 Challenges and Limitations
5.1 Genetic diversity and inbreeding concerns
One of the primary challenges in using quantitative genetics to improve carcass traits in pigs is maintaining genetic diversity while avoiding inbreeding. Inbreeding can lead to a reduction in genetic diversity, which in turn can result in inbreeding depression, negatively affecting traits such as growth, meat quality, and overall health of the pigs. Studies have shown that while heritability estimates for carcass traits can be moderate to high, the genetic correlations between these traits can vary significantly, indicating the complexity of genetic interactions (Miar et al., 2014a; Miar et al., 2014b). For instance, the heritability estimates for carcass traits ranged from 0.22 to 0.63, suggesting that while genetic improvement is possible, careful management of genetic diversity is crucial to avoid negative consequences (Miar et al., 2014b).
Moreover, the genetic correlations between purebred and crossbred pigs can be inconsistent, further complicating breeding programs. The genetic correlations between purebred and crossbred performance (rpc) for various traits have been found to be moderate to high, but with large standard errors, indicating potential issues with the accuracy of selection. This variability underscores the importance of incorporating crossbred performance data into breeding programs to ensure that genetic gains in purebred populations translate effectively to commercial crossbred populations (Esfandyari et al., 2019; Esfandyari et al., 2020).
5.2 Environmental and management factors affecting carcass traits
Environmental and management factors play a significant role in the expression of carcass traits, adding another layer of complexity to genetic improvement programs. Factors such as housing conditions, feed quality, and climate can significantly influence the phenotypic expression of genetic traits. For example, genotype by environment interactions (GxE) have been observed, indicating that pigs raised in different environmental conditions (e.g., tropical vs. temperate climates) may exhibit different genetic correlations for carcass traits (Godinho et al., 2019). This suggests that breeding programs need to account for environmental variability to achieve consistent genetic improvements across different production systems.
Additionally, management practices such as feeding regimes and housing conditions can impact carcass traits. Studies have shown that significant fixed effects such as company, sex, and slaughter batch, as well as covariates like cold carcass weight and slaughter age, need to be considered in genetic models to accurately estimate genetic parameters (Miar et al., 2014; Miar et al., 2014b). These factors can introduce variability that may mask the true genetic potential of the animals, making it essential to standardize management practices as much as possible to achieve reliable genetic evaluations.
5.3 Ethical considerations in genetic selection
Ethical considerations are increasingly important in the context of genetic selection for improved carcass traits in pigs. One major ethical concern is the welfare of the animals. Intensive selection for specific traits, such as increased muscle mass or reduced fat, can lead to unintended negative consequences on animal health and well-being. For instance, selection for increased daily gain and reduced backfat thickness has been associated with potential deterioration in pork quality and increased susceptibility to stress and disease (Miar et al., 2014; Willson et al., 2020). Therefore, it is crucial to balance genetic improvement with the overall welfare of the animals to ensure sustainable and ethical breeding practices.
Another ethical issue is the potential impact on biodiversity. Intensive selection for specific traits can lead to a narrowing of the genetic base, reducing the overall genetic diversity within pig populations. This reduction in diversity can make populations more vulnerable to diseases and environmental changes, posing a risk to long-term sustainability. Ethical breeding programs should aim to maintain genetic diversity while achieving desired improvements in carcass traits. This can be done by incorporating a wide range of genetic lines and avoiding excessive inbreeding (Miar et al., 2014b; Esfandyari et al., 2019; Esfandyari et al., 2020). Additionally, transparent communication with stakeholders about the goals and methods of genetic selection is essential to address ethical concerns and build public trust in genetic improvement programs.
6 Future Directions in Quantitative Genetics for Carcass Traits
6.1 Integrating omics technologies with quantitative genetics
The integration of omics technologies with quantitative genetics holds significant promise for advancing our understanding and improvement of carcass traits in pigs. Omics technologies, including genomics, transcriptomics, proteomics, and metabolomics, provide comprehensive insights into the biological mechanisms underlying phenotypic traits. These technologies enable the construction of regulatory networks that integrate different levels of biological information flow from gene to function, thereby offering a holistic view of the genetic architecture of complex traits (Keurentjes et al., 2008; Verardo et al., 2023). For instance, the AnimalQTLdb project has curated extensive genomic information on quantitative trait loci (QTL) identified in various livestock species, facilitating the translation of genome to phenome research (Verardo et al., 2023).
Moreover, the application of omics technologies in livestock genetics has led to the development of important tools for improving animal production and sustainability. Projects like the Functional Annotation of Animal Genomes (FAANG) have generated datasets to decipher the function of genome segments in multiple species, including pigs (Verardo et al., 2023). These datasets, combined with advanced analytical and statistical techniques, allow for more accurate genetic predictions and better understanding of the genomic background of phenotypic variability. As a result, integrating omics technologies with quantitative genetics can significantly enhance the accuracy and efficiency of breeding programs aimed at improving carcass traits in pigs (Chakraborty et al., 2022).
6.2 Potential of CRISPR and other gene-editing tools
The advent of CRISPR and other gene-editing tools has revolutionized the field of genetics, offering unprecedented opportunities for precise genetic manipulation. CRISPR technology, characterized by its low technological barrier and high efficiency, has been successfully employed in various applications, including the improvement of economically important traits in pigs (Tu et al., 2022; Wang and Doudna, 2023). For example, gene editing has been used to knockout the Myostatin gene to enhance lean meat production and to knock-in the UCP1 gene to improve piglet thermogenesis and survival under cold stress (Tu et al., 2022). These advancements demonstrate the potential of CRISPR to rapidly and precisely alter genes responsible for desirable carcass traits.
Looking forward, the continued development and refinement of CRISPR technology will likely address current challenges such as improving editing accuracy and precision, and enhancing targeted delivery of CRISPR editors (Figure 2) (Wang and Doudna, 2023). Additionally, the integration of CRISPR with other emerging technologies like machine learning and live cell imaging could further expand its applications in both fundamental and applied research. As regulatory frameworks evolve to accommodate these new breeding technologies, the commercialization and global valorization of gene-edited pigs are expected to increase, potentially transforming the pig industry by enabling the production of healthier, more productive animals (Tu et al., 2022).
 Figure 2 New ICT based fertility management model in private dairy farm India as well as abroad |
6.3 Enhancing accuracy and efficiency in genetic prediction
Enhancing the accuracy and efficiency of genetic prediction is crucial for the success of breeding programs aimed at improving carcass traits in pigs. Recent studies have demonstrated the potential of genomic selection to predict carcass quality traits with reasonable accuracy. For instance, a study evaluating the predictive ability of different models for carcass traits in crossbred pigs found that the inclusion of additional records in the training set significantly improved prediction accuracies (Bergamaschi et al., 2019). This highlights the importance of large, well-annotated datasets and robust statistical models in achieving accurate genetic predictions.
Furthermore, the integration of multi-omics data with traditional quantitative genetics approaches can enhance the precision of genetic predictions. By leveraging comprehensive datasets that include genomics, transcriptomics, proteomics, and metabolomics information, researchers can gain a deeper understanding of the genetic basis of complex traits and identify key genetic markers associated with desirable carcass traits (Chakraborty et al., 2022). This multi-faceted approach not only improves the accuracy of breeding value estimates but also accelerates the genetic gain by enabling the selection of superior animals at an early stage of life. As omics technologies continue to advance and become more accessible, their integration with quantitative genetics will play a pivotal role in the future of pig breeding programs (Keurentjes et al., 2008; Verardo et al., 2023).
7 Concluding Remarks
This study of quantitative genetics in improving carcass traits in pigs has revealed several significant insights. Genome-wide association studies (GWAS) and quantitative trait loci (QTL) mapping have identified numerous genetic markers associated with economically important traits such as back fat thickness, meat fat ratio, carcass length, and average daily gain. For instance, pooling F2 designs and resequencing efforts have led to the discovery of over 32 million variants, including 8 million novel ones, and identified significant variant clusters on chromosomes 1, 2, 4, 7, 17, and 18. Additionally, QTL analyses in various pig populations have consistently highlighted significant loci on chromosomes 1, 2, 4, 7, and X, affecting traits like lean content, fat content, and dressing percentage. Heritability estimates for carcass and meat quality traits have been found to be moderate to high, indicating the potential for genetic improvement through selective breeding.
The findings from these studies have profound implications for pig breeding and meat production. The identification of specific genetic markers and QTLs allows for the development of marker-assisted selection (MAS) programs, which can accelerate genetic improvement in pig populations. For example, the discovery of high-impact variants and candidate genes such as BMP2 can be directly utilized in breeding programs to enhance traits like back fat thickness and carcass length. The moderate to high heritability estimates for traits such as trimmed ham weight and longissimus dorsi muscle area suggest that these traits can be effectively improved through both traditional and genomic selection methods. Furthermore, the genetic correlations between performance traits and meat quality traits indicate that it is possible to select for increased growth rates without adversely affecting meat quality, thus optimizing both production efficiency and product quality.
While significant progress has been made in understanding the genetic basis of carcass traits in pigs, several areas warrant further research. Future studies should focus on fine-mapping the identified QTL regions to pinpoint the exact genes and causal variants responsible for the observed phenotypic variations. Additionally, integrating multi-omics approaches, such as transcriptomics and proteomics, could provide deeper insights into the molecular mechanisms underlying these traits. There is also a need for more extensive validation of identified markers across diverse pig populations to ensure their broad applicability in different breeding contexts. Finally, exploring the potential of gene editing technologies, such as CRISPR/Cas9, could open new avenues for directly modifying key genetic loci to achieve desired carcass traits more efficiently. By addressing these areas, future research can further enhance the precision and effectiveness of genetic improvement programs in pig breeding.
Acknowledgements
Thank you to the peer reviewers for their constructive feedback on the initial draft of this manuscript.
Conflict of Interest Disclosure
Author affirms that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Arruda M.P., Lipka A.E., Brown P.J., Krill A.M., Thurber C., Brown-Guedira G., Dong Y., Foresman B.J., and Kolb F., 2016, Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.), Molecular Breeding, 36: 1-11.
https://doi.org/10.1007/s11032-016-0508-5
Bergamaschi M., Maltecca C., Schwab C., Fix J., and Tiezzi F., 2019, 213 Genomic selection of carcass quality traits in crossbred pigs using a reference population, Journal of Animal Science, 97(Suppl 3): 41.
https://doi.org/10.1093/jas/skz258.082
Biermann A.D.M., Yin T., von Borstel U.U.K., Rübesam K., Kuhn B., and König S., 2015, From phenotyping towards breeding strategies: using in vivo indicator traits and genetic markers to improve meat quality in an endangered pig breed, Animal, 9(6): 919-927.
https://doi.org/10.1017/S1751731115000166
PMID: 25690016
Budhlakoti N., Kushwaha A.K., Rai A., Chaturvedi K.K., Kumar A., Pradhan A.K., Kumar U., Kumar R.R., Juliana P., Mishra D., and Kumar S., 2022, Genomic selection: a tool for accelerating the efficiency of molecular breeding for development of climate-resilient crops, Frontiers in Genetics, 13: 832153.
https://doi.org/10.3389/fgene.2022.832153
PMID: 35222548 PMCID: PMC8864149
Chakraborty D., Sharma N., Kour S., Sodhi S.S., Gupta M., Lee S.J., and Son Y.K., 2022, Applications of omics technology for livestock selection and improvement, Frontiers in Genetics, 13.
https://doi.org/10.3389/fgene.2022.774113
PMID: 35719396 PMCID: PMC9204716
Čepica S., Zambonelli P., Weisz F., Bigi M., Knoll A., Vykoukalová Z., Masopust M., Gallo M., Buttazzoni L., and Davoli R., 2013, Association mapping of quantitative trait loci for carcass and meat quality traits at the central part of chromosome 2 in Italian large white pigs, Meat science, 95(2): 368-375.
https://doi.org/10.1016/j.meatsci.2013.05.002
PMID: 23747631
Duarte D.A.S., Fortes M.R.S., Duarte M.R., Guimarães S.E.F., Verardo L.L., Veroneze R., Ribeiro A.M.F., Lopes P., Resende M., and Silva F.F., 2017, Genome-wide association studies, meta-analyses and derived gene network for meat quality and carcass traits in pigs, Animal Production Science, 58(6): 1100-1108.
https://doi.org/10.1071/AN16018
Duthie C., Simm G., Doeschl-Wilson A., Kalm E., Knap P.W., and Roehe R., 2010, Epistatic analysis of carcass characteristics in pigs reveals genomic interactions between quantitative trait loci attributable to additive and dominance genetic effects, Journal of Animal Science, 88(7): 2219-2234.
https://doi.org/10.2527/jas.2009-2266
Edwards D.B., Ernst C.W, Raney N.E., Doumit M.E., Hoge M.D., and Bates R.O., 2008, Quantitative trait locus mapping in an F2 Duroc x Pietrain resource population: II, Carcass and meat quality traits, Journal of animal science, 86(2): 254-266.
https://doi.org/10.2527/JAS.2006-626
Esfandyari H., Thekkoot D., Kemp R., Plastow G., and Dekkers J., 2020, Genetic parameters and purebred-crossbred genetic correlations for growth, meat quality, and carcass traits in pigs, Journal of Animal Science, 98(12): skaa379.
https://doi.org/10.1093/jas/skaa379
PMID: 33325519 PMCID: PMC7755176
Esfandyari H., Thekkoot D., Kemp R., Plastow G., and Dekkers J., 2019, PSIII-13 genetic parameters and the purebred–crossbred genetic correlation for growth, carcass, and meat quality traits in pigs, Journal of Animal Science, 98(12): skaa379.
https://doi.org/10.1093/JAS/SKZ122.290
PMID: 33325519 PMCID: PMC7755176
Falker-Gieske C., Blaj I., Preuss S., Bennewitz J., Thaller G., and Tetens J., 2019, GWAS for meat and carcass traits using imputed sequence level genotypes in pooled F2-designs in pigs, G3: Genes|Genomes|Genetics, 9(9): 2823-2834.
https://doi.org/10.1534/g3.119.400452
PMID: 31296617 PMCID: PMC6723123
Godinho R., Godinho R., Bergsma R., Silva F., Sevillano C., Knol E.F., Komen H., Guimarães S.E.F., Lopes M.S., and Bastiaansen J.W.M., 2019, Genetic correlations between growth performance and carcass traits of purebred and crossbred pigs raised in tropical and temperate climates, Journal of Animal Science, 97(9): 3648-3657.
https://doi.org/10.1093/jas/skz229
PMID: 31278865 PMCID: PMC6735805
Heffner E.L., Sorrells M.E., and Jannink J.L., 2009, Genomic selection for crop improvement, Crop Science, 49(1): 1-12.
https://doi.org/10.2135/CROPSCI2008.08.0512
Heffner E., Lorenz A., Jannink J., and Sorrells M., 2010, Plant Breeding with genomic selection: gain per unit time and cost, Crop Science, 50(5): 1681-1690.
https://doi.org/10.2135/CROPSCI2009.11.0662
Jiang Z.H., Michal J.J., Chen J., Daniels T., Kunej T., Garcia M., Gaskins C., Busboom J., Alexander L., Jr R., and MacNeil M., 2009, Discovery of novel genetic networks associated with 19 economically important traits in beef cattle, International Journal of Biological Sciences, 5: 528-542.
https://doi.org/10.7150/IJBS.5.528
Keurentjes J., Koornneef M., and Vreugdenhil D., 2008, Quantitative genetics in the age of omics, Current opinion in plant biology, 11(2): 123-128.
https://doi.org/10.1016/j.pbi.2008.01.006
Merrick L., Herr A., Sandhu K., Lozada D., and Carter A., 2022, Optimizing plant breeding programs for genomic selection, Agronomy, 12(3): 714.
https://doi.org/10.20944/preprints202202.0048.v1
Meuwissen T., Hayes B., and Goddard M., 2016, Genomic selection: A paradigm shift in animal breeding, Animal Frontiers, 6(1): 6-14.
https://doi.org/10.2527/AF.2016-0002
Miar Y., Plastow G., Bruce H., Moore S., Manafiazar G., Kemp R., Charagu P., Huisman A., van Haandel B., Zhang C.Y., Mckay R., and Wang Z.Q., 2014a, Genetic and phenotypic correlations between performance traits with meat quality and carcass characteristics in commercial crossbred pigs, PLoS ONE, 9(10): e110105.
https://doi.org/10.1371/journal.pone.0110105
PMID: 25350845 PMCID: PMC4211683
Miar Y., Plastow G., Moore S., Manafiazar G., Charagu P., Kemp R., Haandel B., Huisman A., Zhang C., McKay R., Bruce H., and Wang Z., 2014b, Genetic and phenotypic parameters for carcass and meat quality traits in commercial crossbred pigs, Journal of animal science, 92(7): 2869-2884.
https://doi.org/10.2527/jas.2014-7685
Milan D., Bidanel J.P., Iannuccelli N., Riquet J., Amigues Y., Gruand J., Roy P.L., Renard C., and Chevalet C., 2002, Detection of quantitative trait loci for carcass composition traits in pigs, Genetics, Selection, Evolution : GSE, 34(6): 705-728.
https://doi.org/10.1186/1297-9686-34-6-705
Soares M.H., Rodrigues G., Júnior D., da Silva C.B., Costa T., Duarte M., and Saraiva A., 2022, Performance, carcass traits, pork quality and expression of genes related to intramuscular fat metabolism of two diverse genetic lines of pigs, Foods, 11(15): 2280.
https://doi.org/10.3390/foods11152280
PMID: 35954050 PMCID: PMC9368243
Tu C.F., Chuang C.K., and Yang T.S., 2022, The application of new breeding technology based on gene editing in pig industry-a review, Animal Bioscience, 35: 791-803.
https://doi.org/10.5713/ab.21.0390
Peñagaricano F., Valente B., Steibel J., Bates R., Ernst C., Khatib H., and Rosa G., 2015, Searching for causal networks involving latent variables in complex traits: Application to growth, carcass, and meat quality traits in pigs, Journal of animal science, 93(10): 4617-4623.
https://doi.org/10.2527/jas.2015-9213
Polasik D., Tyra M., Żak G., and Terman A., 2018, An analysis of MYH7 single nucleotide polymorphism (g.7:75667956G>A) in relation to growth and carcass traits in pigs, Journal of Animal and Feed Sciences, 27(4): 335-340.
https://doi.org/10.22358/jafs/98929/2018
Verardo L.L., Brito L., Carolino N., and Magalhães A., 2023, Editorial: omics applied to livestock genetics, Frontiers in Genetics, 14: 1155611.
https://doi.org/10.3389/fgene.2023.1155611
PMID: 36873944 PMCID: PMC9978907
Velez-Irizary, D., Casiro, S., Daza, K., Bates, R., Raney, N., Steibel, J., and Ernst, C., 2019, 33 young scholar presentation: expression quantitative trait loci and allele-specific expression exhibiting joint association with polygenic trait phenotypes in pigs, Journal of Animal Science, 97(Supplement_2): 19.
https://doi.org/10.1093/JAS/SKZ122.035
Vidal O., Noguera J.L., Amills M., Varona L., Gil M., Jiménez N., Dávalos G., Folch J., and Sánchez A., 2005, Identification of carcass and meat quality quantitative trait loci in a Landrace pig population selected for growth and leanness, Journal of Animal Science, 83(2): 293-300.
https://doi.org/10.2527/2005.832293X
PMID: 15644499
Wang J.Y., and Doudna J.A., 2023, CRISPR technology: A decade of genome editing is only the beginning, Science, 379(6629): eadd8643.
https://doi.org/10.1126/science.add8643
PMID: 36656942
Willson H.E., de Oliveira H.R., Schinckel A.P., Grossi D., and Brito L., 2020, Estimation of genetic parameters for pork quality, novel carcass, primal-cut and growth traits in duroc pigs, Animals, 10(5): 779.
https://doi.org/10.3390/ani10050779
PMID: 32365996 PMCID: PMC7278482
Willson H., Oliveira H., Schinckel A., and Grossi D., 2019, PSVIII-37 Estimation of genetic parameters for novel meat quality and carcass traits in duroc pigs, Journal of Animal Science, 97(Suppl 3): 265.
https://doi.org/10.1093/jas/skz258.539
Yang R.F., Guo X., Zhu D., Tan C., Bian C., Ren J., Huang Z., Zhao Y., Cai G.Y., Liu D., Wu Z., Wang Y., Li N., and Hu, X., 2021, Accelerated deciphering of the genetic architecture of agricultural economic traits in pigs using a low-coverage whole-genome sequencing strategy, GigaScience, 10(7): giab048.
https://doi.org/10.1093/gigascience/giab048
PMID: 34282453 PMCID: PMC8290195
Zhang Y.F., Zhang J.J., Gong H., Cui L., Zhang W., Ma J., Chen C., Ai H., Xiao S.J., Huang L.S., and Yang B., 2019, Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations, Meat science, 150: 47-55.
https://doi.org/10.1016/j.meatsci.2018.12.008
PMID: 30584983

. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Qiqi Zhou
. Shiqiang Huang
Related articles
. Pig carcass traits
. Quantitative genetics
. Breeding strategies
. Marker-assisted selection
. Genomic selection
Tools
. Post a comment


